About

Omics Expert & Computational Biologist
Passionate about microbiome research and multi-omics
- Date of birth: 6 July 1990
- Email: menia.gavr@gmail.com
- Phone: +31 620607736
- City: Wageningen, The Netherlands
- Degree: PhD
- Expertise: Omics, NGS,Microbiome Research
- ORCID: https://orcid.org/0000-0003-0204-2223
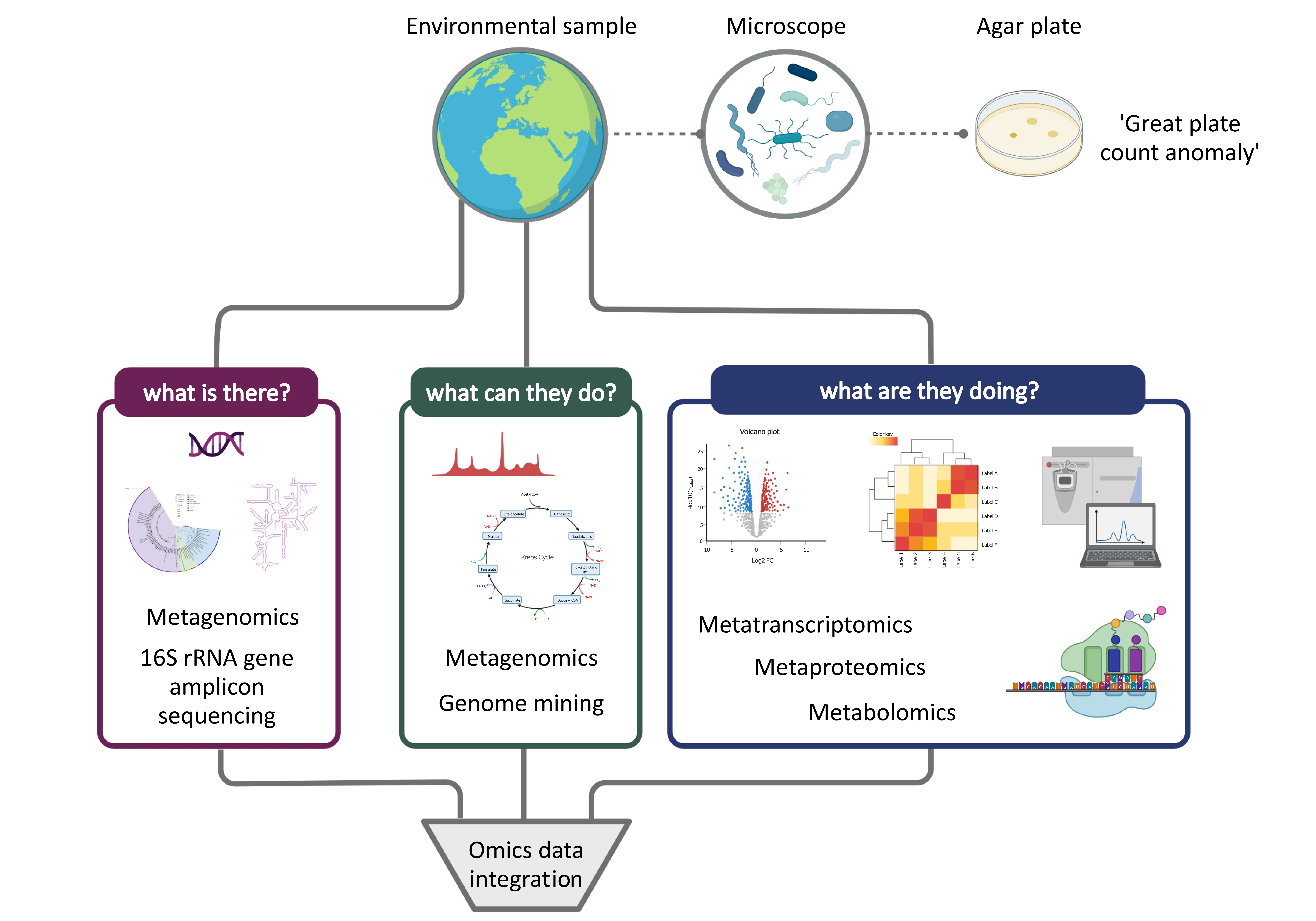
As a computational biologist, I specialize in omics and microbiome research, with a strong background in data analysis and interpretation. Proficient in utilizing advanced computational tools and bioinformatics techniques to uncover valuable insights from complex biological datasets. Accomplished in executing analytical pipelines, as well as collaborating with interdisciplinary teams to drive impactful research in the fields of (meta)genomics, (meta)transcriptomics, proteomics, and microbiome analysis. Harnessing data-driven approaches to enhance our understanding of microbial communities and their interactions with their host environments is my passion!
Facts
My expertise lies in handling and analyzing large-scale microbiome datasets, including data preprocessing, quality control, statistical analysis and integration from diverse sources. My enthusiasm stems from the potential to contribute to groundbreaking discoveries in (micro)biology, ultimately advancing our knowledge of the intricacies of life at a molecular level.
Publications in scientific journals, preprint servers, international symposia and conferences
Check them out!Citations in peer-reviewed scientific journals
EU Projects and Consortia supported my research
Years of experience working in multidisciplinary teams in various universities and research institutes
Skills
NGS Data Analysis
- Preprocessing and quality control of raw data
- Sequence alignment and mapping
- Assembly of (meta)genomes and (meta)transcriptomes
- Metagenome binning and comparative genomics
- Taxonomic assignment and phylogenomics
- Functional annotation and Metabolism reconstruction
- Database manipulation
- Proteomics data analysis
- Biological interpretation of large-scale microbiome datasets
Unix/Linux Operation
- File manipulation and navigation
- Advanced shell scripting (Bash)
- Package and environment management (e.g., Conda)
- Utilization of bioinformatic tools and pipelines
- High-Performance and Cloud Computing (AWS)
Programming
- R: Data manipulation (dplyr, tidyr), statistical analysis (Bioconductor), data visualization (ggplot2, plotly), and data reporting (R Markdown)
- Python: Data manipulation and analysis (pandas), data visualization (seaborn, matplotlib), statistical analysis (scipy), and numerical computing (numpy)
Other software
- Illustration tools (Adobe Illustrator, Adobe Photoshop, Inkscape)
- Version control with Github and Git
- Code editing with VS code, Submlime Text and Notepad++
- Microsoft office suite
Scientific Communication
- Technical Report & Research Article writing
- Project Report writing for international & national research collaborations
- Present research findings at meetings, symposia & conferences
- Peer-reviewed articles for high-impact factor scientific journals
Analytical & Interpersonal skills
- Teamwork
- Proactivity
- Critical thinking
- Problem solving
- Pattern recognition
- Attention to detail
- Time management
Resume
Professional Experience
Postdoctoral Researcher
2022 - Present
Wageningen, The Netherlands
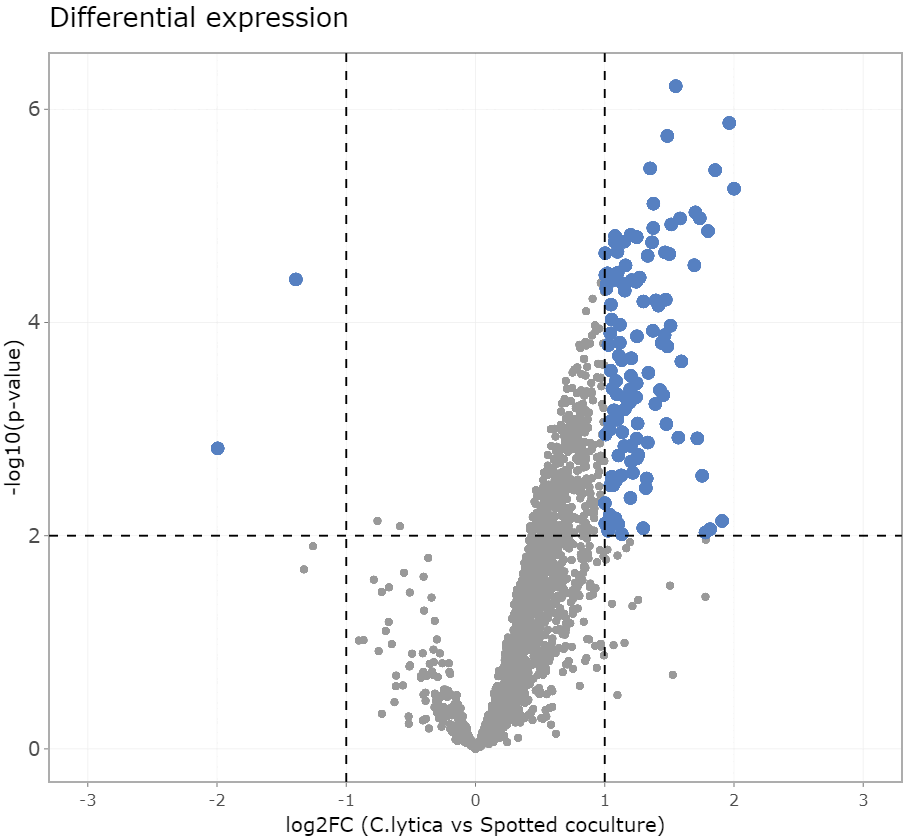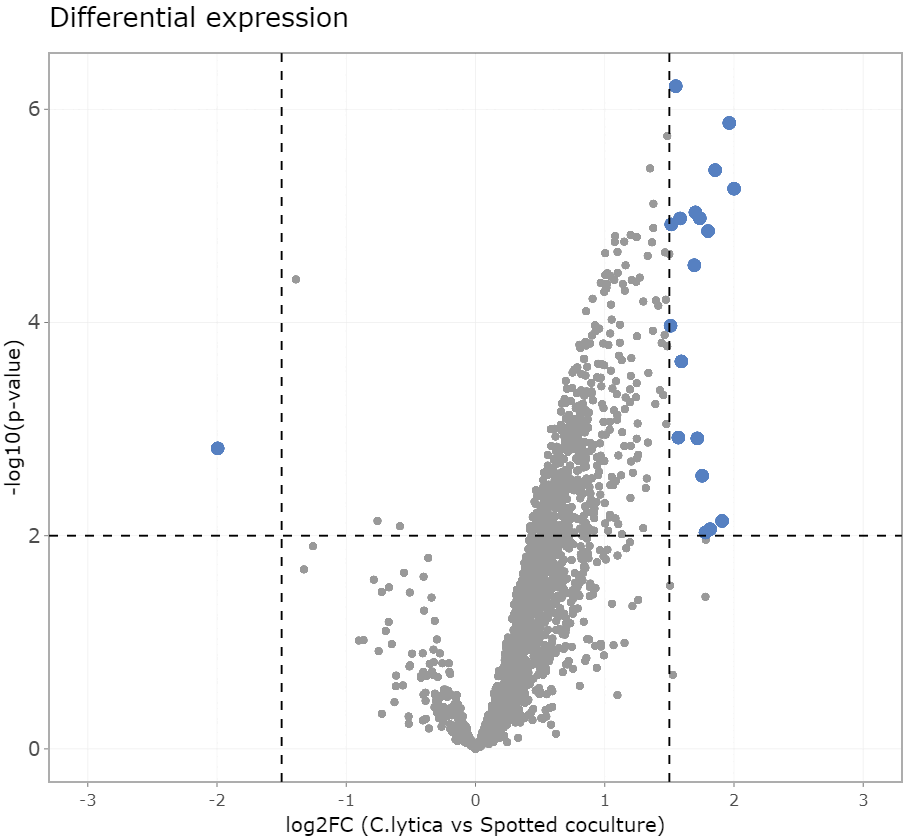
- Led and conducted multi-omics projects focusing on microbial and host-microbe interactions in human and animal health, nutrition and biotechnological applications
- Analysed high-throughput sequencing data employing advanced bioinformatic tools and workflows
- Facilitated collaborations across disciplines to ensure successful translation of microbiome research
- Mentored PhD students in research question formulation, experimental design and data analysis
- Contributed to high-impact publications, peer-reviewed journals and presented findings at scientific conferences
- Drafted and contributed to grant proposals and project applications
- Data management according to data science best practices
- Tutorial development on NGS data analysis
Marie Skłodowska-Curie Early Stage Researcher (ESR)
2017 - 2022
Wageningen, The Netherlands
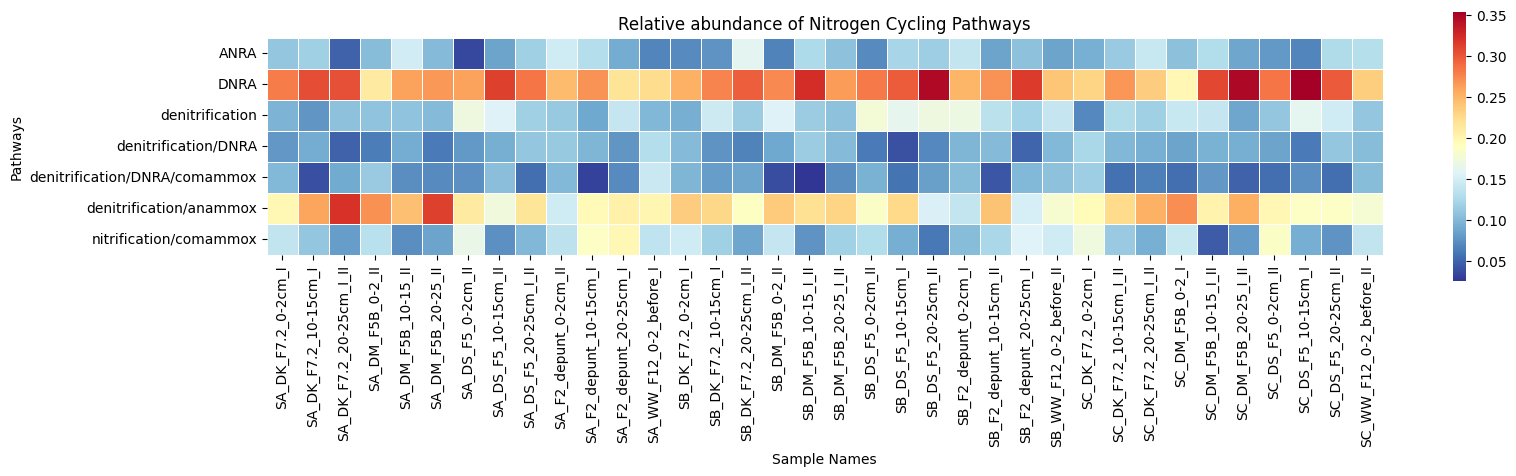
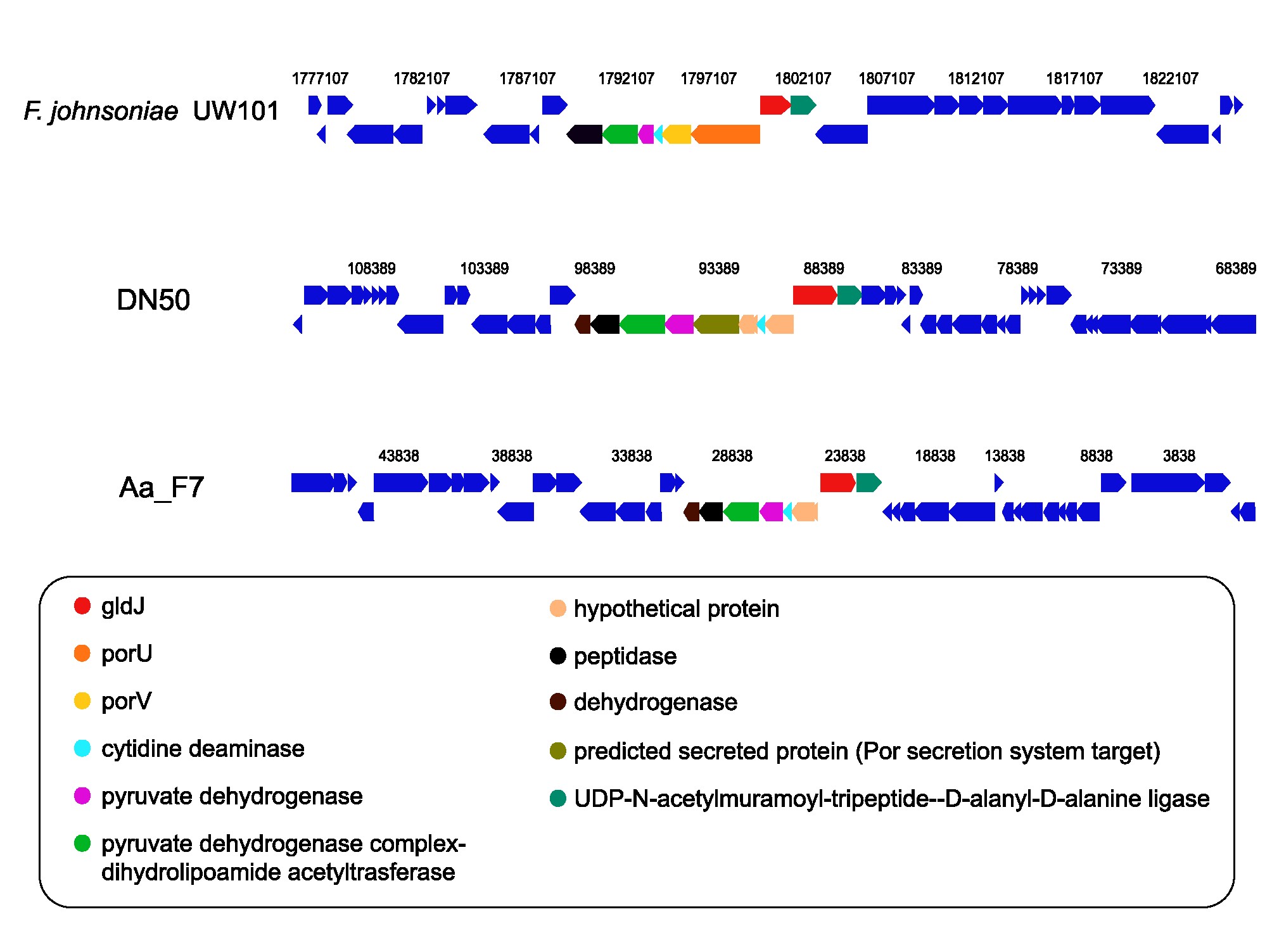
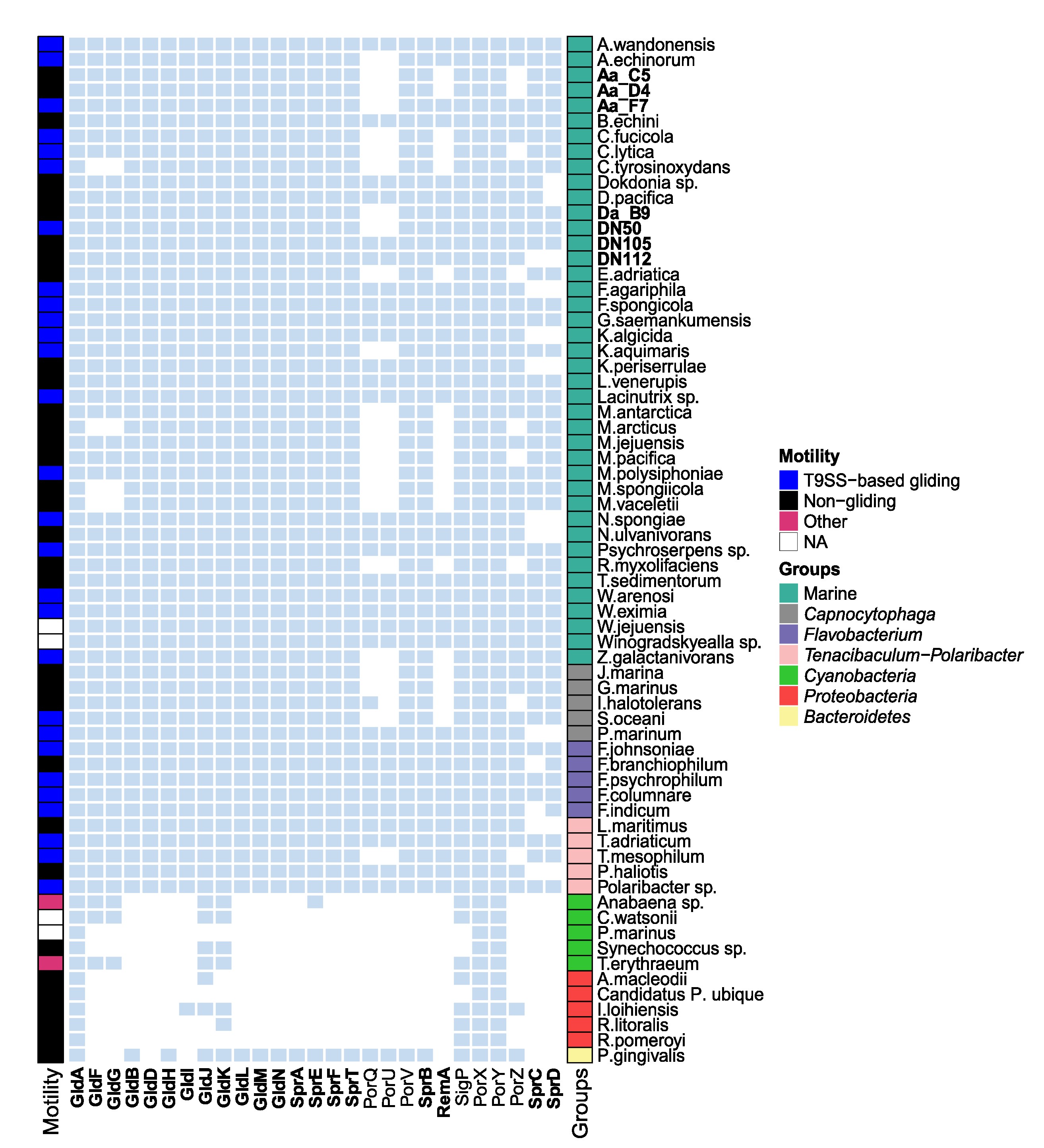
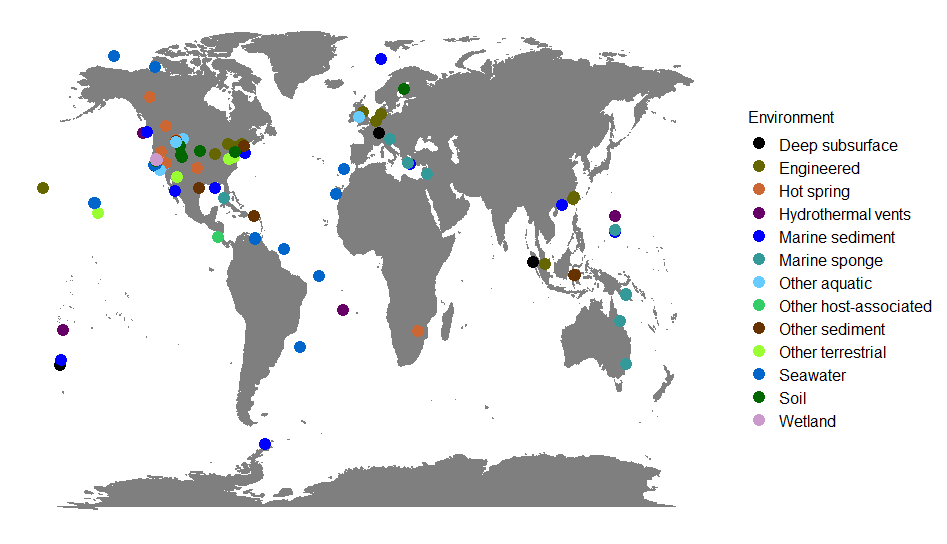
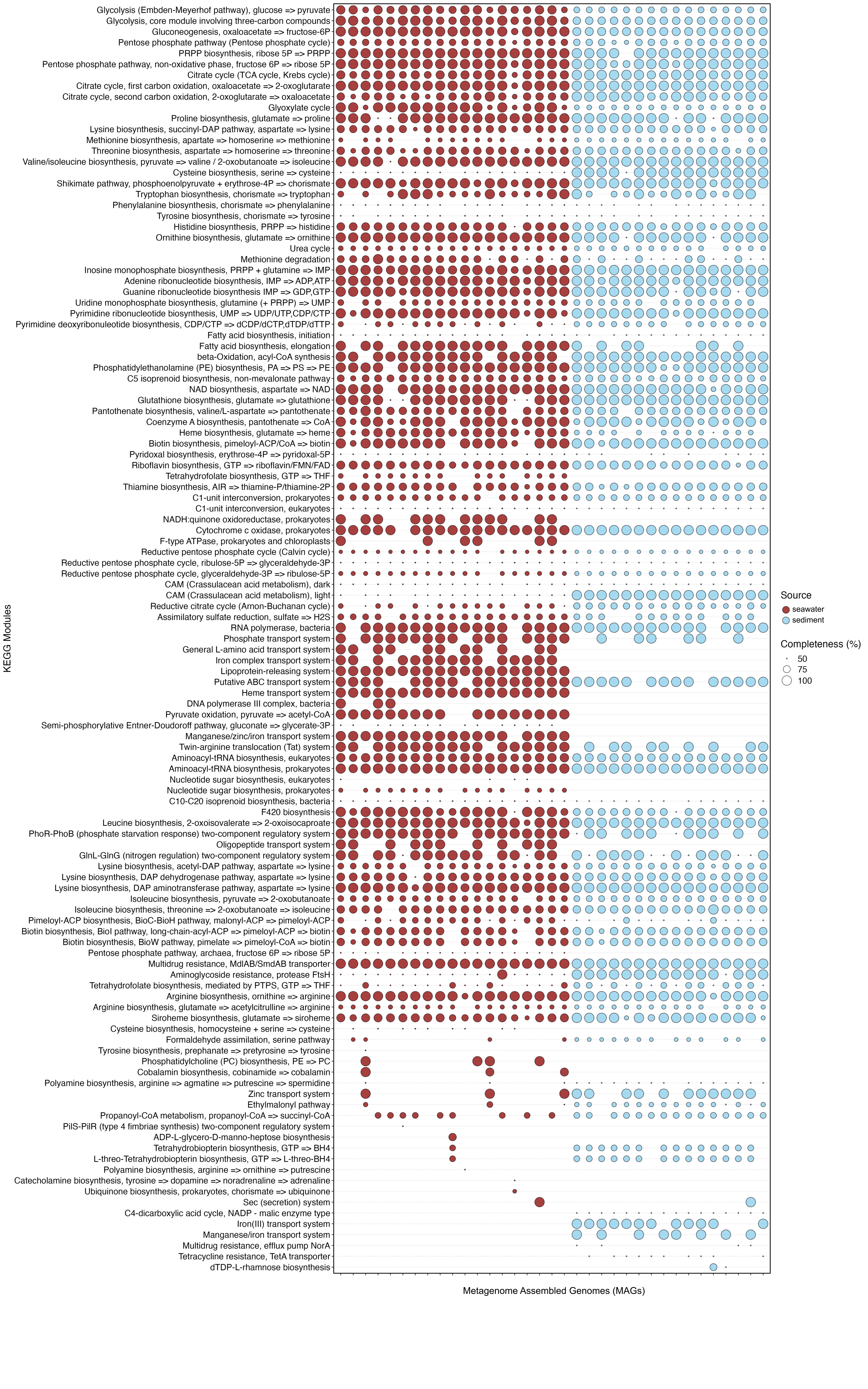
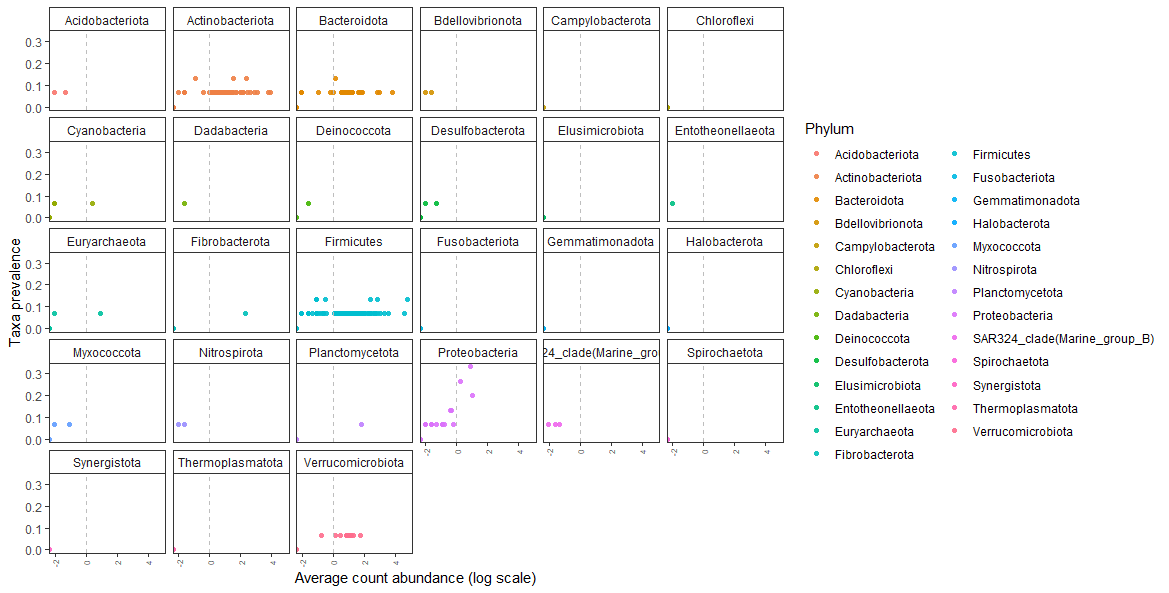
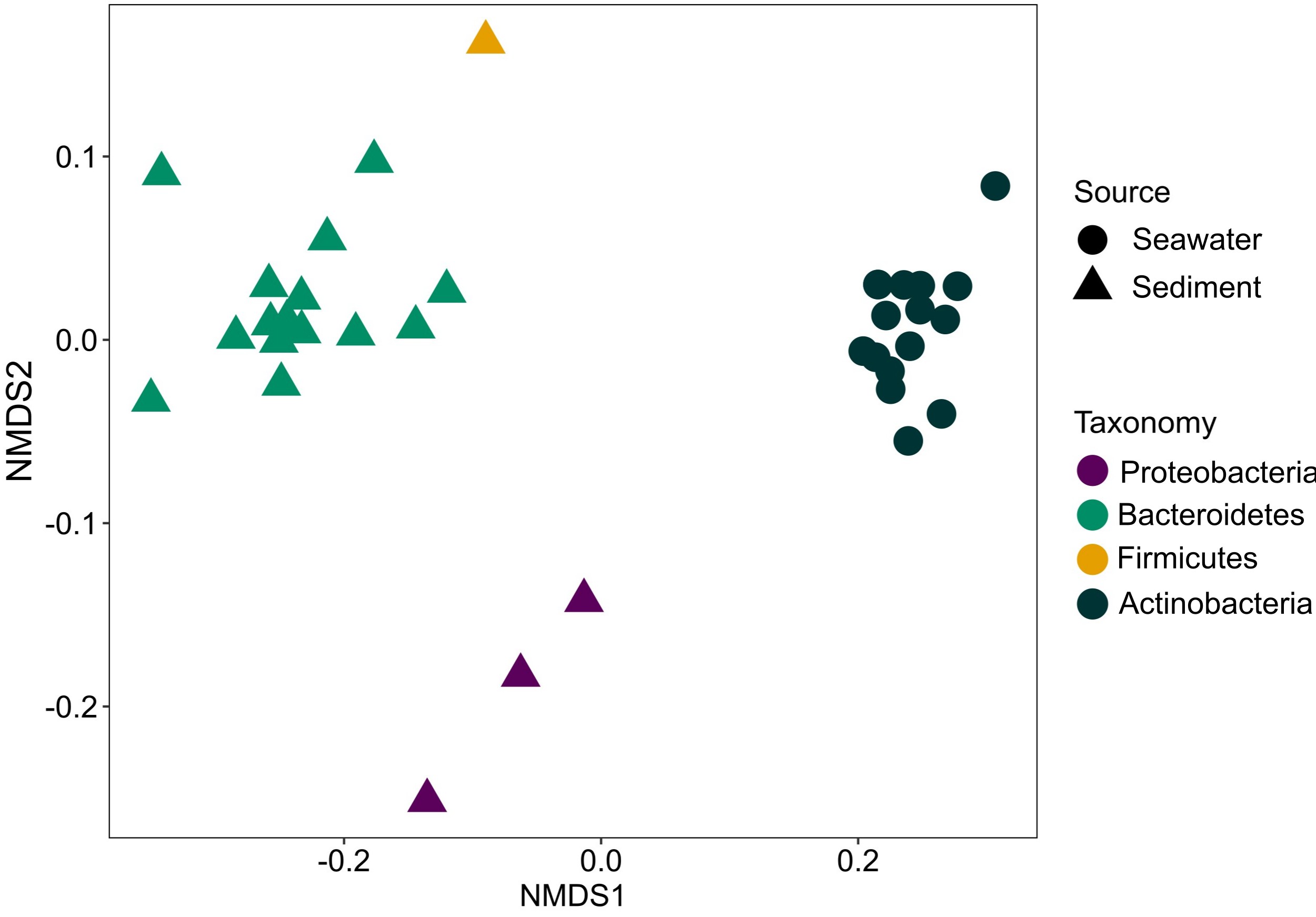
- Conducted metagenomic analysis to unravel host-microbiome interactions
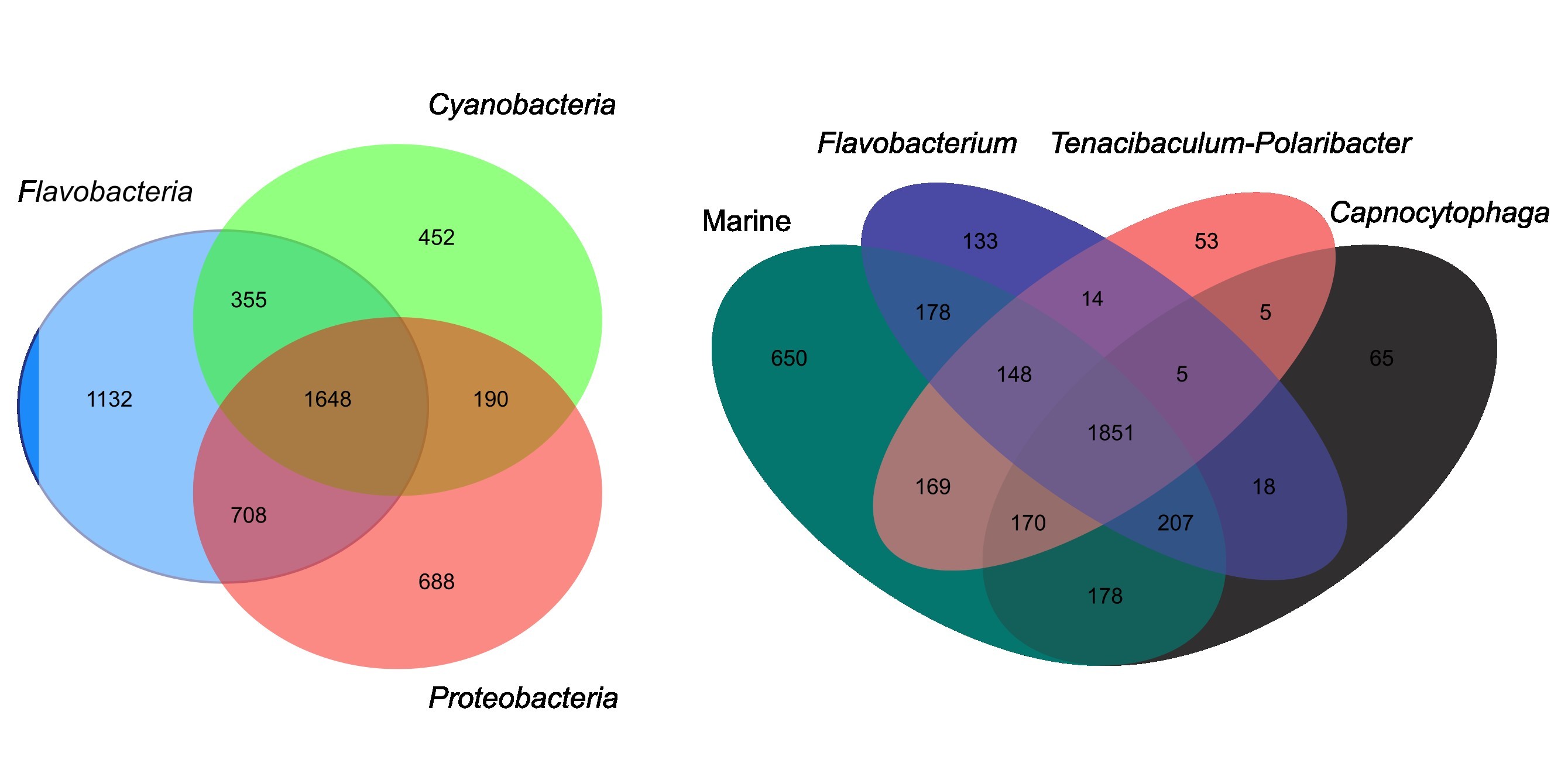
- Utilized various bioinformatic tools and pipelines for metagenome binning and comparative genomics
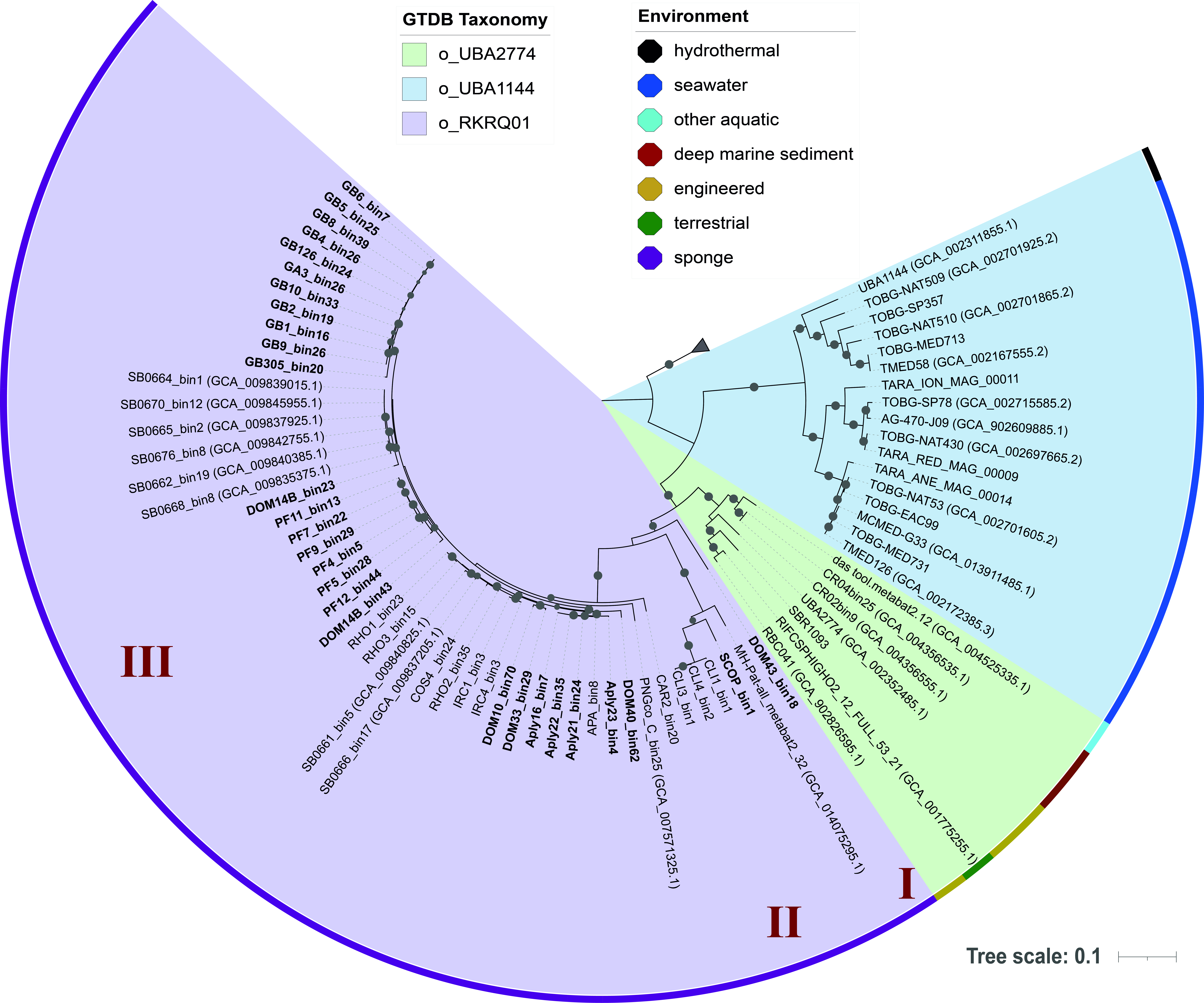
- Performed taxonomic profiling to reconstruct the phylogeny of host-associated microbiomes
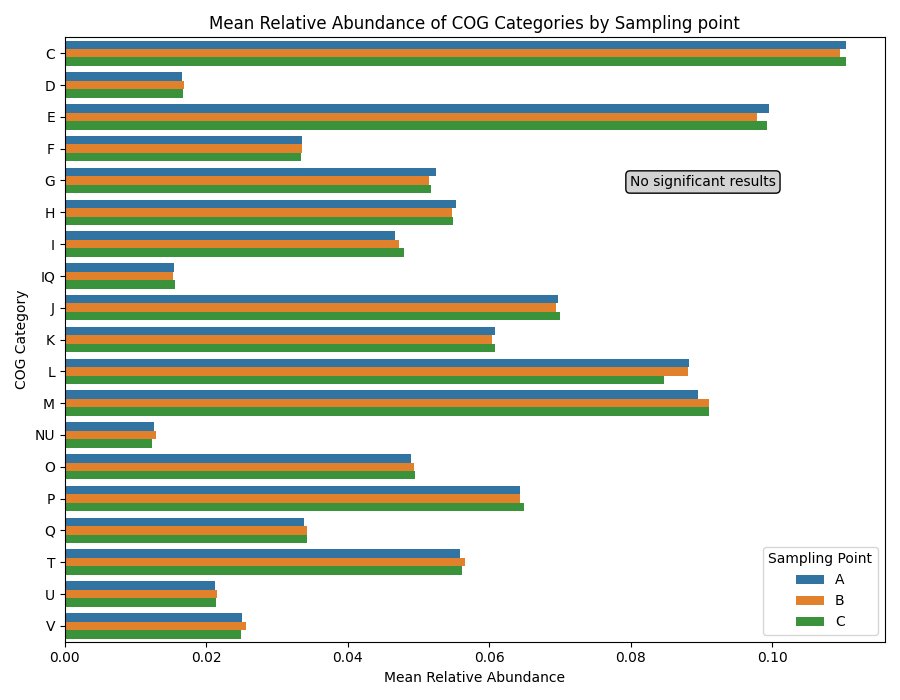
- Assessed the functional potential by mining metagenomes and single genomes for metabolic pathways, carbohydrate-active enzymes and biosynthetic gene clusters
- Published analyses and findings in peer-reviewed scientific journals
- Presented results in consortia meetings, symposia and international conferences
Education
PhD in Microbiology
2017 - 2022
Laboratory of Microbiology, Wageningen University & Research, The Netherlands
Master of Science in Environmental Biology
2014 - 2016
University of Crete, Greece
Minor: Management of Marine Biological Resources
Bachelor of Science in Biology
2008 - 2013
Aristotle University of Thessaloniki, Greece
List of Projects
Human Gut Microbiome Profiling in High-Risk Diabetes
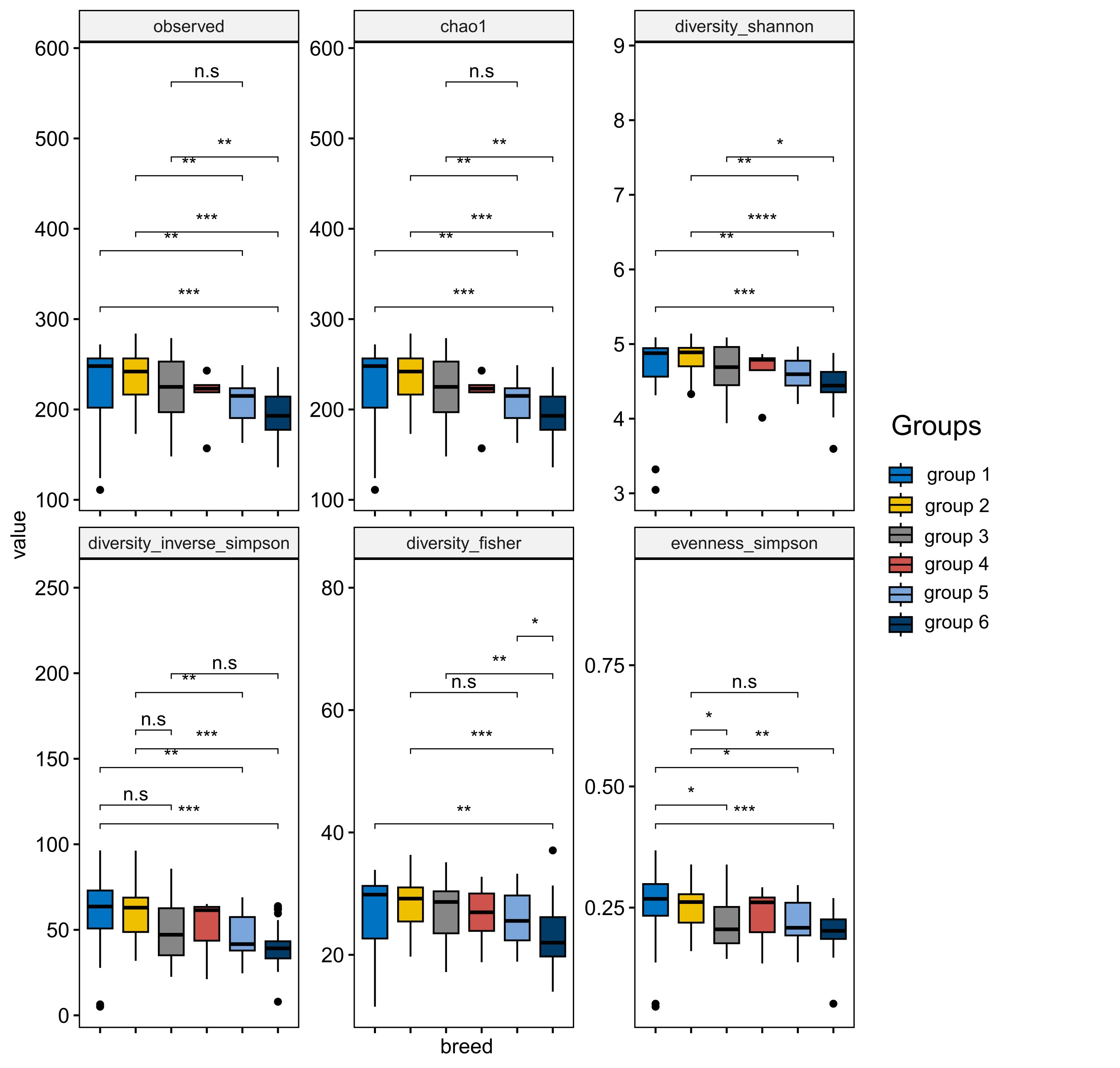
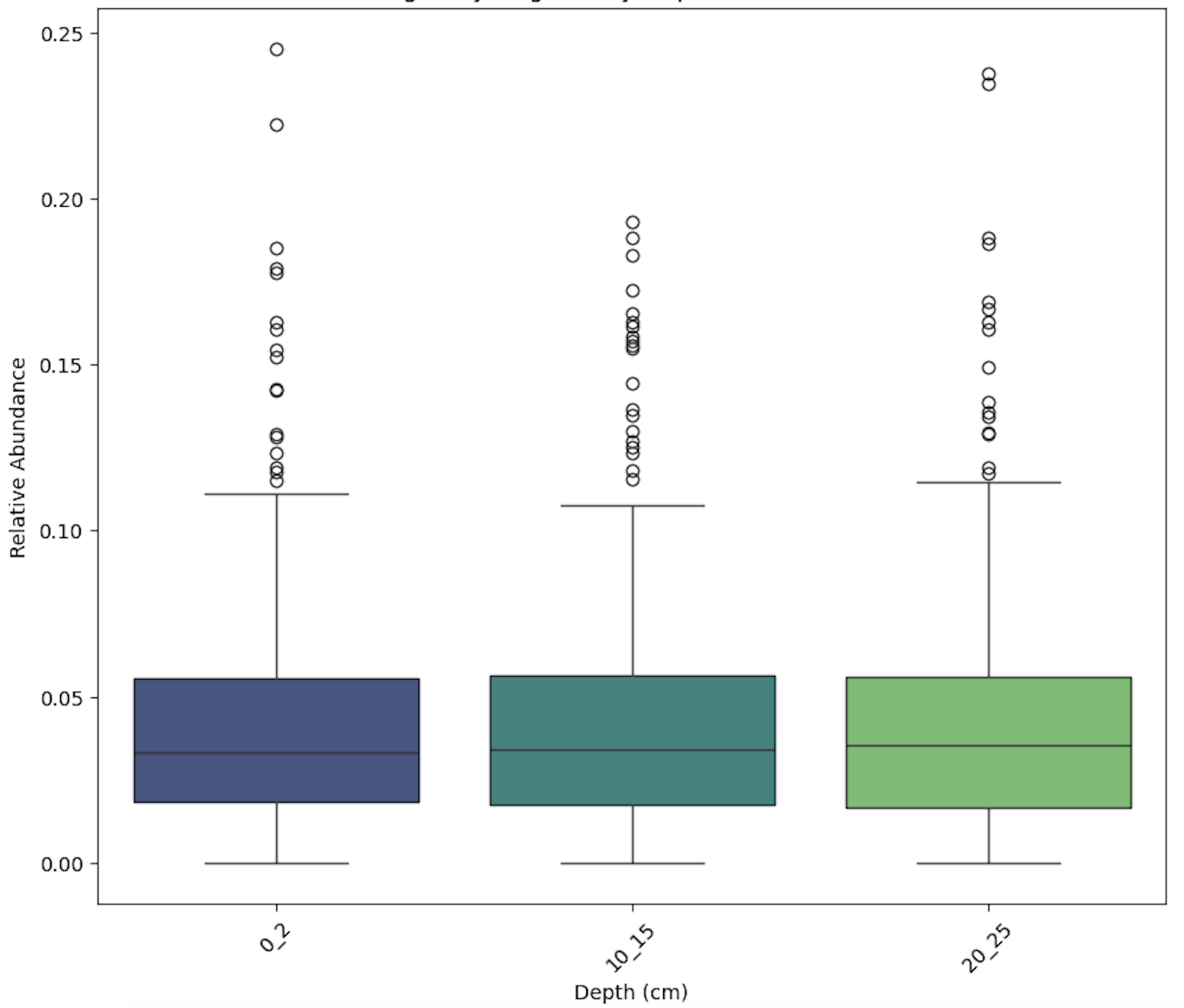
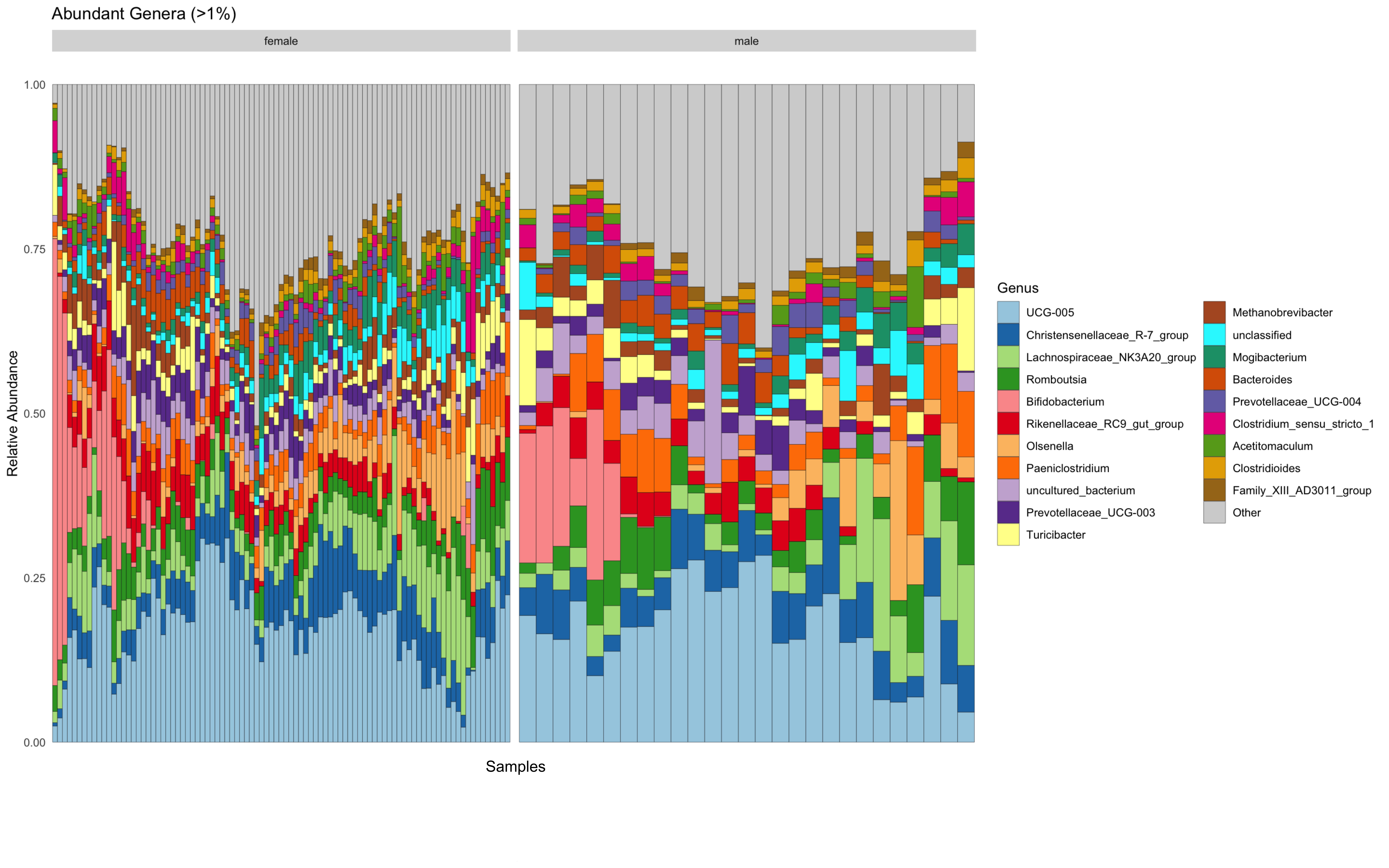
- Analysed metagenomes to study microbial community structure and functional potential in individuals with type 2 diabetes risk, focusing on dietary interventions with fibre intake
Impact of Prebiotic Oligosaccharides on Pediatric Gut Health
- Developed statistical analysis plans for two clinical trials. Performed statistical analysis of clinical data and analysis of 16S rRNA sequencing data to evaluate the effect of prebiotics on gut microbiota in children with functional constipation
Functional Analysis of Microbial Fermentation of Dietary Fibers
- Conducted metatranscriptomic analysis to assess the impact of dietary fibres on microbial diversity and metabolic activity, providing insights into human gut health
Dietary Supplement Effects on Gut Microbiota in Osteoarthritis Patients
- Analysed gut microbiota composition and functional profiles using 16S rRNA and metagenomic sequencing to understand the role of dietary supplements in modulating gut health
Contact
You can contact me via email or social media!
Location:
Wageningen, The Netherlands
Email:
menia.gavr@gmail.com